Kneron End to End Simulator v0.24.0
This project allows users to perform image inference using Kneron's built in simulator. We encourage users to use simply use the kneron_inference function to perform the tests on your inputs.
Python API Inference
This is a standalone feature that allows the user to perform inference given a model and some inputs. This is separate from the E2E Simulator itself. For details, you can take a look at /workspace/E2E_Simulator/python_flow/kneron_inference.py.
Necessary items
There are only three items that you need to prepare to run the inference function. Everything else is optional.
- preprocessed input: list of NumPy arrays in channel first format (matches ONNX). Before 0.21.0, this took in channel last format.
- input_names: list of strings indicating the inputs to the model
- model file: depending on what kind of model you want to run, but it will be one of NEF, ONNX, and BIE file.
Inputs
def kneron_inference(pre_results: List[npt.ArrayLike], input_names: List[str],
nef_file: str = "", onnx_file: str = "", bie_file: str = "",
model_id: Optional[int] = None, data_type: str = "float",
reordering: Optional[List[str]] = None,
platform: int = 520) -> List[npt.ArrayLike]:pre_results: same aspreprocessed inputmentioned aboveinput_names: list of strings indicating the inputs to the model- order of names should match the order of inputs in
pre_results nef_file/onnx_file/bie_file: string path to your input model file- only one of these will be used, if they are all specified, priority is NEF -> ONNX -> BIE
model_id: integer ID of model to run inference- only necessary if NEF file with multiple models is used
data_type: string data format that you would like the output returned asfloatorfixedreordering: list of strings specifying the output order- this is mainly to match the output order desired for the postprocess
- each item in the list should be a name of an output node in the model
platform: integer platform to be used- only needed for BIE/NEF models
- supported options:
520,530,630,720,730
Output
Output will be a list of NumPy arrays in ONNX shape format. It will be in the order specified by reordering; if reordering is not specified, it will be in the default order provided by the model.
Usage
Prepare a preprocess function and postprocess function. Then, simply call the kneron_inference function using the results of your preprocess function and input model file to get inference results. Then, use those inference results as input into your postprocess function.
Example
For a detailed example on how to call the kneron_inference function, please explore this YOLO example.
Setting up C preprocess/postprocess
- Compile the C/C++ functions into a shared library (.so). Be sure to add
extern "C"to any C++ functions you intend to directly call in the Python flow. -
Import the shared library into whichever module needs it using the standard ctypes module:
MY_LIB = ctypes.CDLL("path/to/my_lib.so") -
Define any C/C++ structures that are needed for parameters as classes in Python. These classes must extend
ctypes.Structureand store the structure fields in the_fields_variable. This is a list of tuples where the first item is the name of the variable, and the second item is the type of that variable. The names and order must be defined exactly as in the C code. - Define a wrapper to the C/C++ function you wish to call. You need to specify three items: the function name as defined in the C code, the input argument types, and the result argument types.
- For preprocess, make sure that the image data needed for preprocessing can be passed from Python into your C function and that the resulting preprocessed data can be passed from C into Python. When passed into the kneron_inference function, the data should be in ONNX shape.
- Similarly, for the postprocess, make sure that the inference output can be passed from Python into your C function and that the resulting postprocess data can be passed from C into Python.
There are some conversion functions for your convenience under python_flow/utils/utils.py.
Use convert_binary_to_numpy to get a NumPy array from an input binary image. Use convert_pre_numpy_to_rgba to dump a NumPy array into RGBA binary used for CSIM.
FAQ
1. How can I display bounding box results on my image?
There is a display function in /workspace/E2E_Simulator/eeeeeepython_flow/utils/utils.py that can be called. When using this function, the list of bounding boxes input will need to follow a specific format. Each box in the list must have the following values to display the correct box on the image: (x, y, w, h, score, class).
2. How do I convert the CSIM fixed point inference results into floating point results?
The inference output when using CSIM 520/720 are fixed point values in a text file. By default, the simulator will convert those fixed point values into floating point NumPy arrays. You can set the data_type parameter to the kneron_inference function to get the inference results as fixed point values. The conversion from fixed to floating point values is as follows:
float_val = fixed_val / scale / (2 ^ radix)Through CSIM, every output node has a corresponding scale and radix value. For 520, this is stored in a file called radix_scale_info.txt in the same directory as the output values. For 720, this is stored in a file called dma2seq.info per output node in the same directory as the output values.
3. What are the memory layouts for the inference results of CSIM 520 and 720?
The memory layout for the output node data after CSIM inference is different between 520 and 720. For 520, the data values are in (h, c, w_align) format, where w_align is the width aligned to the nearest 16 bytes. For 720, the data values are in (c, h, w_align) format, where w_align is the width aligned to the nearest 16 bytes.
Let's look at an example where c = 4, h = 12, and w = 12. Indexing starts at 0 for this example.
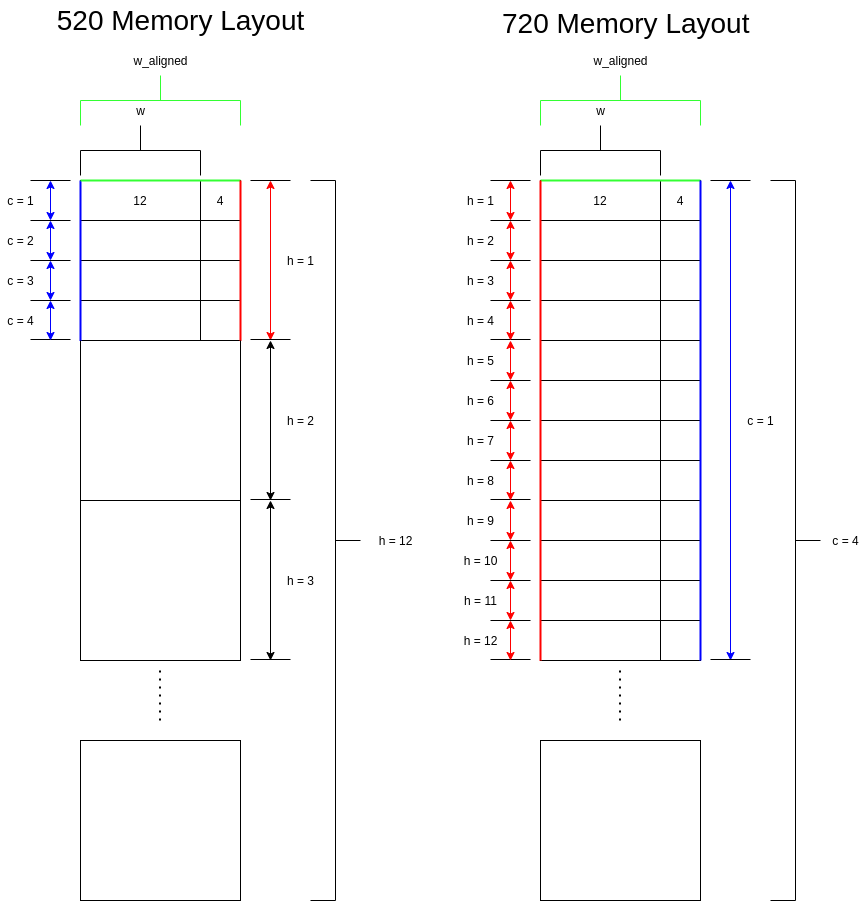
Memory layouts
For both 520 and 720, we can see that the first 12 bytes of data correspond to the width dimension. The next 4 bytes will then be filled with whatever data is in that memory after inference, since the width dimension is aligned to the nearest 16 bytes. The following is where we see the difference in layouts between 520 and 720: for 520, the channel dimension is next, but for 720, the height dimension is next.
For 520, the next 16 bytes of data correspond to channel of 1, while for 720, the next 16 bytes of data correspond to height of 1. Let's take the 20th byte of data as an example. For 520, this would correspond to c = 1, w = 4, and h = 0. However, for 720, this would correspond to c = 0, w = 4, and h = 1.
To recap, for 520, width increments first, up to the aligned 16 byte width. Channel increments next, and height increments last. For 720, width increments first, also up to the aligned 16 byte width. Height increments next, and channel increments last.